
The sampled sequences agreed qualitatively with an experimental library of Tiam1-binding peptides.
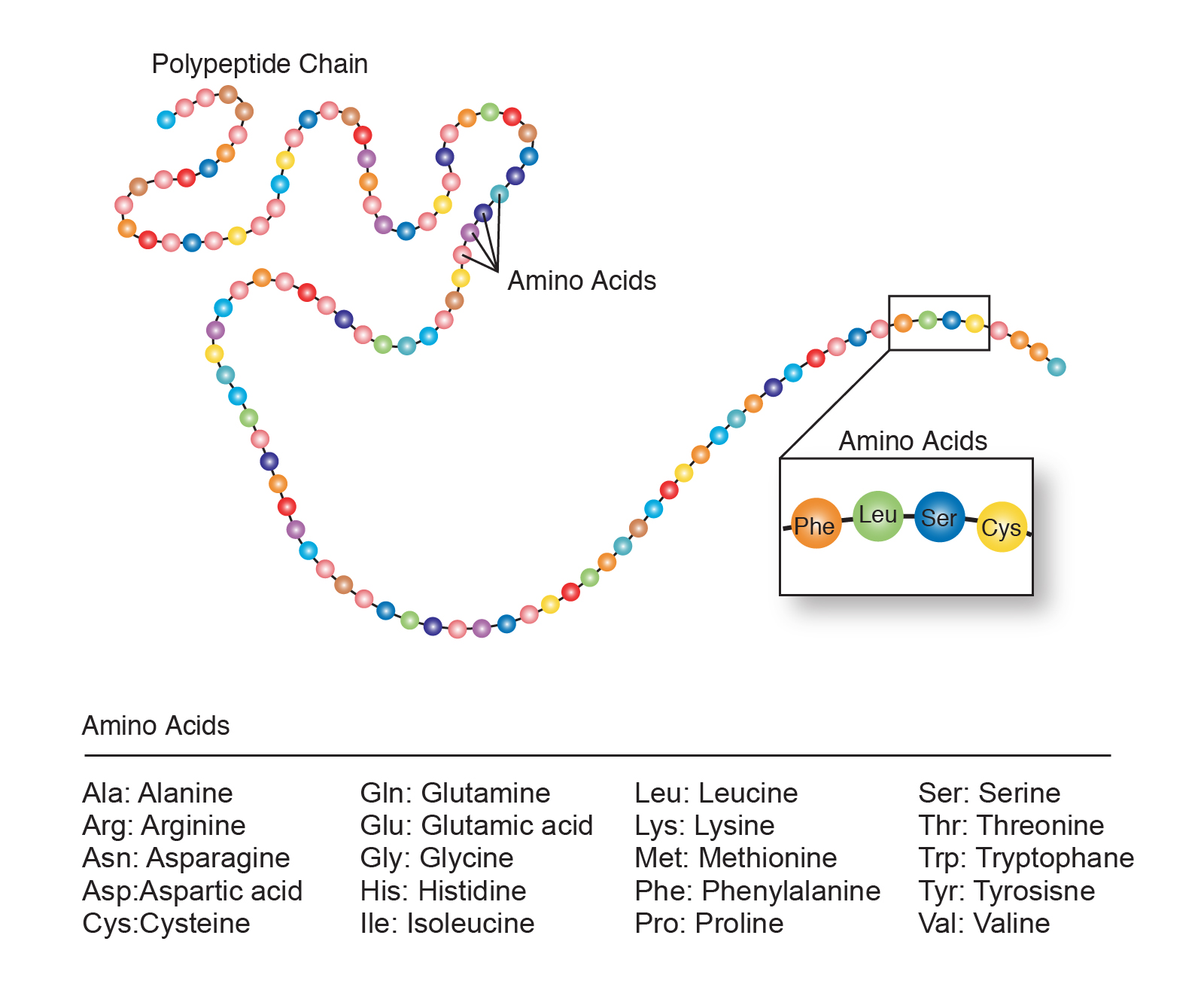
AMINO ACID SEQUENCE FREE
The relative binding free energies agreed qualitatively with values from experiment.
96% of the theoretical sequence space was sampled. Almost 75 000 peptide variants were processed in two simulations of 10 9 steps each that used 1 CPU hour on a desktop machine. Four peptide positions were allowed to mutate freely. We used the method to design peptides that bind to the PDZ domain of the Tiam1 signaling protein and could serve as inhibitors of its activity. They include the use of Replica Exchange Monte Carlo and landscape flattening for both the unbound and bound peptides. We propose several improvements that allow still more efficient sampling and can address larger design problems. The method allows the binding free energy to be used directly as the design criterion. Far more efficient than enumeration is a recent Monte Carlo approach that adaptively flattens the energy landscape in sequence space of the unbound peptide and provides formally exact binding free energy differences. This complex problem is usually addressed with either simple heuristic scoring or expensive sequence enumeration schemes.
For the high throughput design of protein:peptide binding, one must explore a vast space of amino acid sequences in search of low binding free energies.


 0 kommentar(er)
0 kommentar(er)
